DMREF Specific Highlights
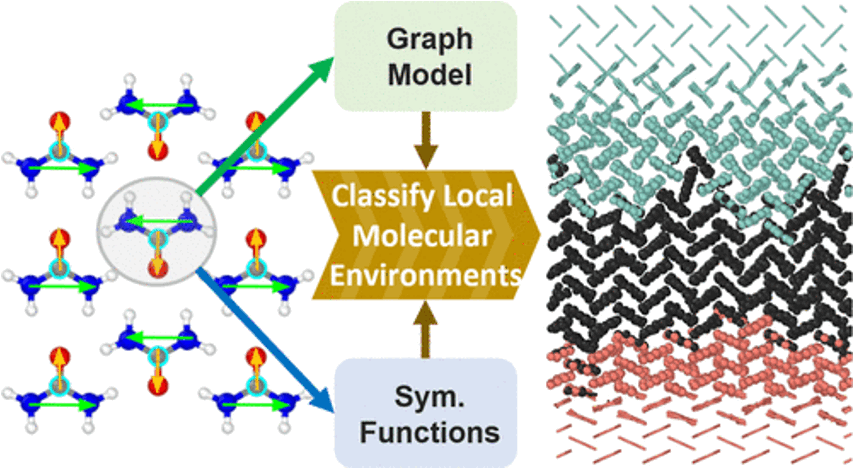
Machine Learning Classification of Local Environments in Molecular Crystals
9/23/2025 | Mark Tuckerman and Jutta Rogal (New York University)
This paper introduces two machine-learning approaches to classify local environments in molecular crystal polymorphs. One uses an atomistic graph convolutional network with molecule-wide aggregation for per-molecule environment classification; the other uses symmetry-function descriptors with a point-vector representation to encode positions and orientations. Both achieve very high accuracy on urea and nicotinamide polymorphs and can analyze dynamical trajectories of nanocrystals and solid–solid interfaces. The methods apply to many molecules and topologies, advancing the study of condensed-matter phenomena.

AI-Driven Defect Engineering for Advanced Thermoelectric Materials
8/22/2025 | Mingda Li (MIT)
A recent review looks at how artificial intelligence (AI) and machine learning (ML) are improving the design of thermoelectric materials, which convert waste heat into electricity. The authors discuss how advanced AI techniques help manage the challenges of defects and optimize material properties. They highlight new strategies for engineering defects and creating materials with specific traits. The review emphasizes the potential of AI in discovering innovative thermoelectric materials and exploring sustainable practices for the future.

Design and Discovery of Organic Semiconductors Aided by Machine Learning
8/19/2025 | Chad Risko (University of Kentucky)
Recent advancements in using artificial intelligence (AI), particularly machine learning (ML), are being applied to improve the design and discovery of organic semiconductors (OSCs). These materials have unique properties that are useful in energy, electronics, and display technologies. The research reviews how ML can help predict material properties and find new molecular building blocks, making it easier to navigate the complex relationships between synthesis, structure, and functionality in OSCs.

Big-data Explorations for Small-molecule, Ionic Isolation Lattices (SMILES)
8/13/2025 | A. Flood and K Raghavachari (Indiana Univ.) and S. Pamidighantam (Georgia Tech.)
A campaign was launched to gather data on molecular dyes in existing literature, focusing on those compatible with SMILES design rules. Researchers identified 47 established fluorophores, including some from previously unused families. Their analysis showed that many cationic dyes come from the cyanine or rhodamine families and are under 2 nm in size, but only 10% of these dyes had complete electrochemical data. Scientists are encouraged to share more data to improve the design process for new molecular dyes using generative AI.
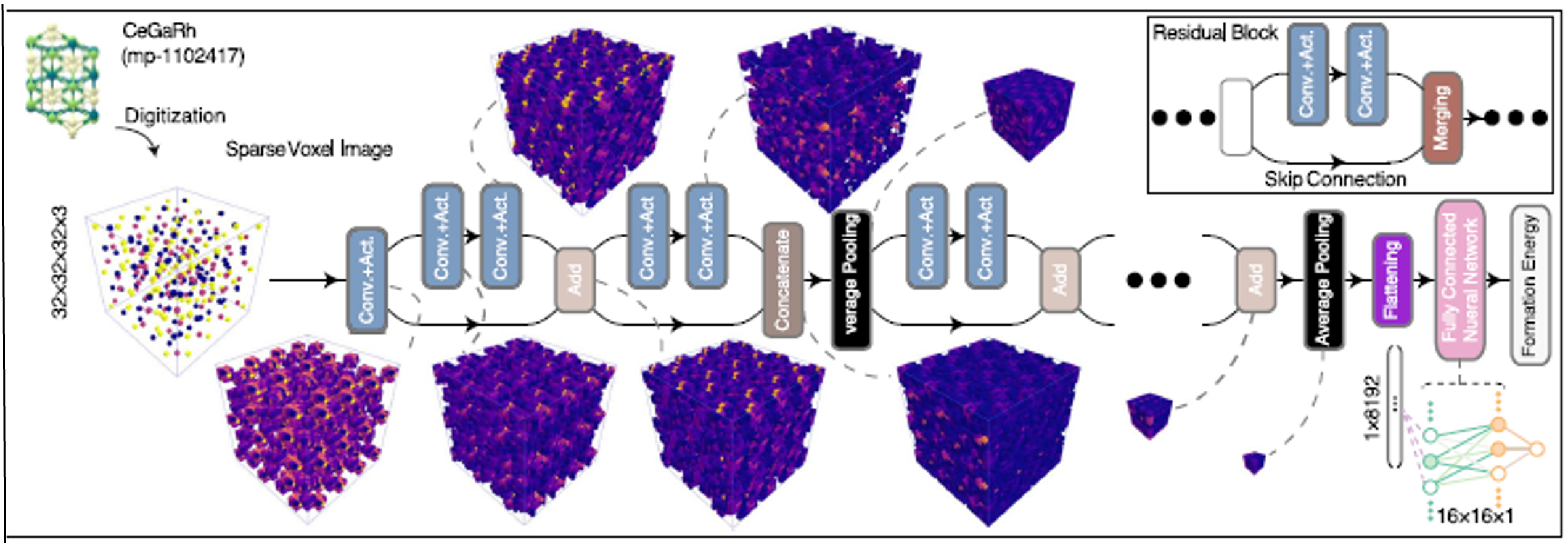
Voxel Image Representation Learning of Crystalline Materials for Formation Energy Prediction
6/20/2025 | Sara Kadkhodaei (University of Illinois-Chicago)
This study highlights the use of visual image representation and deep convolutional neural networks (CNNs) to predict material properties, specifically the formation energy of crystalline materials. By focusing solely on the visual aspects, the researchers found that important material characteristics can be learned without extra physical data. This groundbreaking approach showcases the potential of using computer vision in materials science and opens the door for future advancements, including the possibility of designing materials with specific properties.
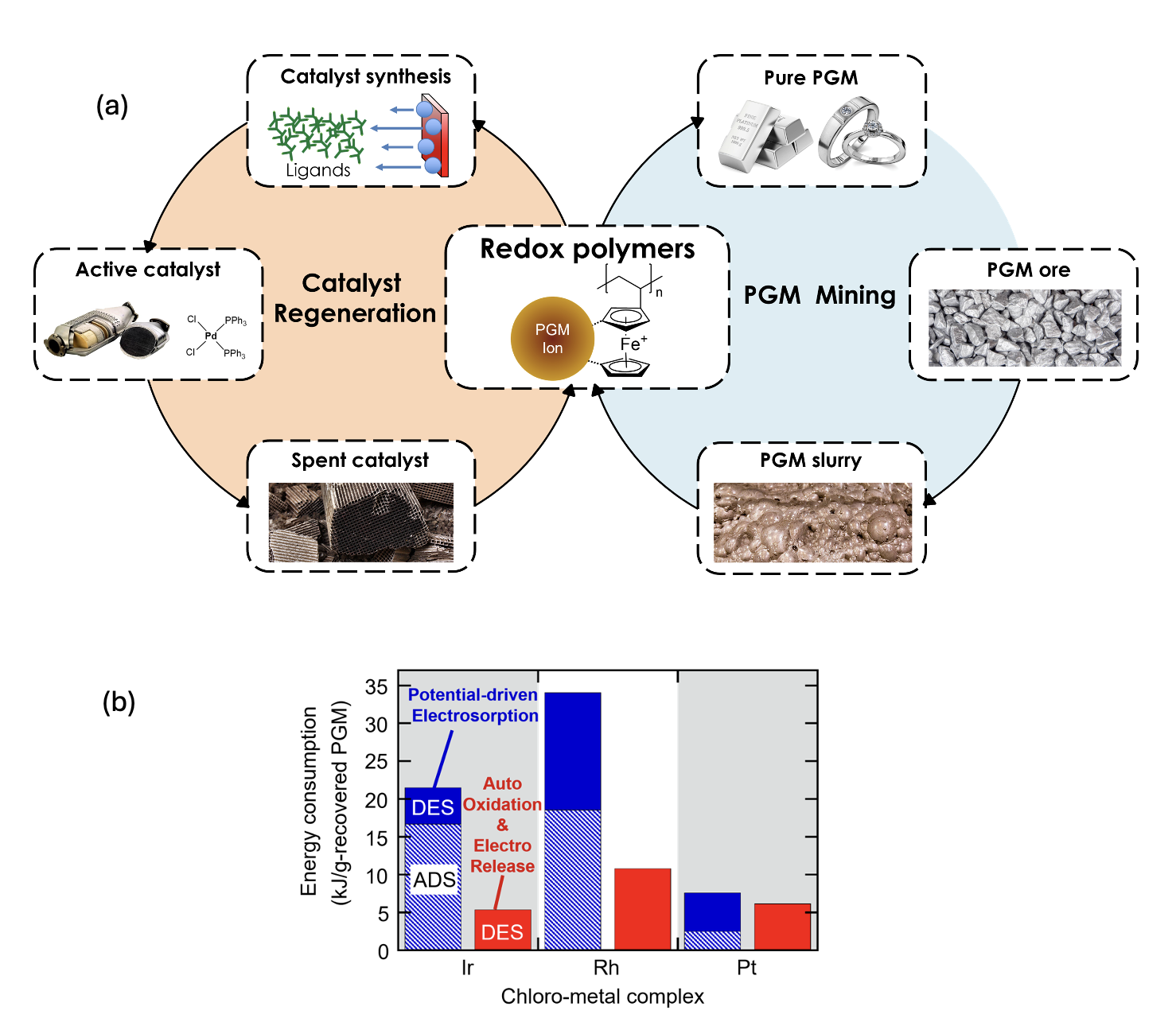
Rational design of redox-responsive materials for critical element separations
11/21/2024 | Xiao Su (University of Illinois Urbana-Champaign)
To develop a new redox material with higher PGM uptake and selectivity, we must understand the effect of metallopolymers for PGMs selectivity more deeply. We plan to combine the spectroscopy and chemical calculation to understand the binding mechanism and the structure effect of redox polymers for PGMs separations. Moreover, to make the recovery system more economical, our goal is to synthesize new redox metallopolymers and immobilized ligands based on the results of computational simulations in an iterative fashion.
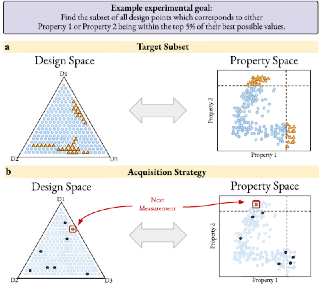
Targeted Materials Discovery using Bayesian Algorithm Execution
8/2/2024 | F. H. da Jornada (Stanford)
Here, a framework is presented that captures experimental goals through straightforward user-defined filtering algorithms. This approach is demonstrated on datasets for TiO2 nanoparticle synthesis and magnetic materials characterization and shows that these methods are significantly more efficient than state-of-the-art approaches.

Machine-learning-driven Expansion of 1D van der Waals Materials Space
8/2/2024 | L. Bartels and A. Balandin (U. CA-Riverside) E. Reed and F. H. da Jornada (Stanford)
Machine-learning techniques were utilized to discover 1D vdW compositions that have not yet been synthesized. This model identified MoI3, a material which was experimentally confirmed to exist with wire-like subcomponents and exotic magnetic properties.
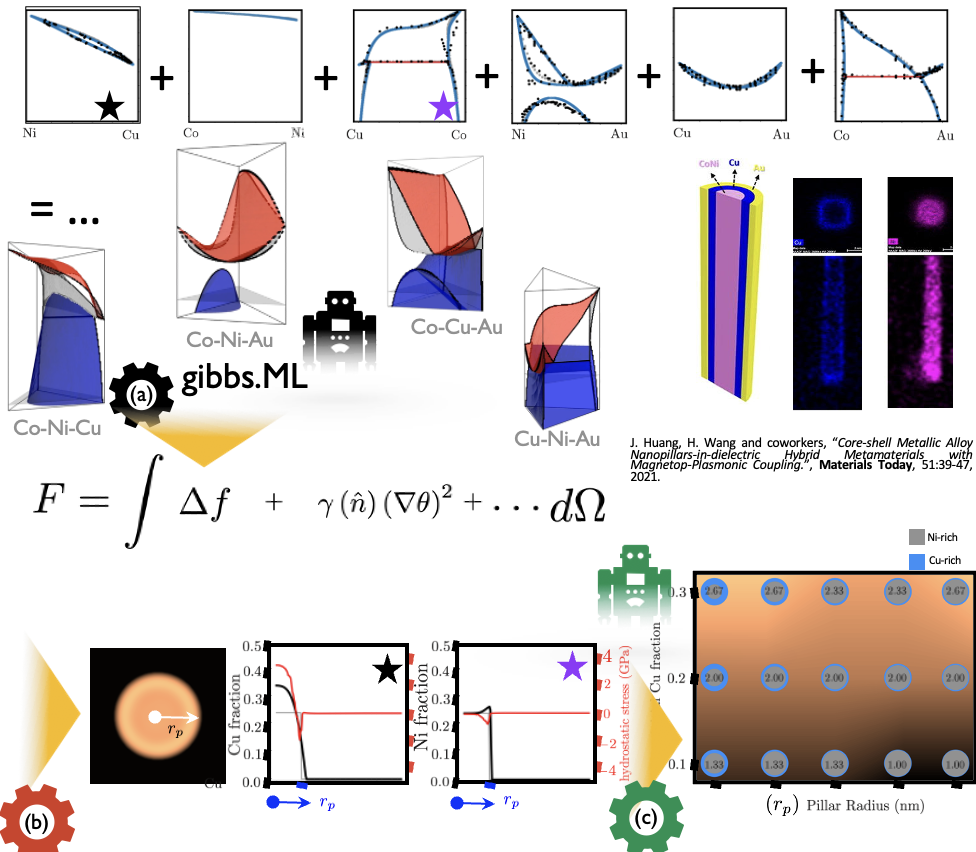
ML-based, Data-driven Infrastructure to Accelerate the Modeling and Simulation of Nanostructures
6/24/2024 | Haiyan Wang (Purdue University)
ML-powered data analytics framework, incorporating the effect available experimental thermodynamic data into phase field models as generated by gibbs.ML in quaternary Ni-Cu-Co-Au-based nanopillars. The generated model allows to streamline the simulation of the segregation dynamics during processing, and generate maps as a function of composition and morphology.
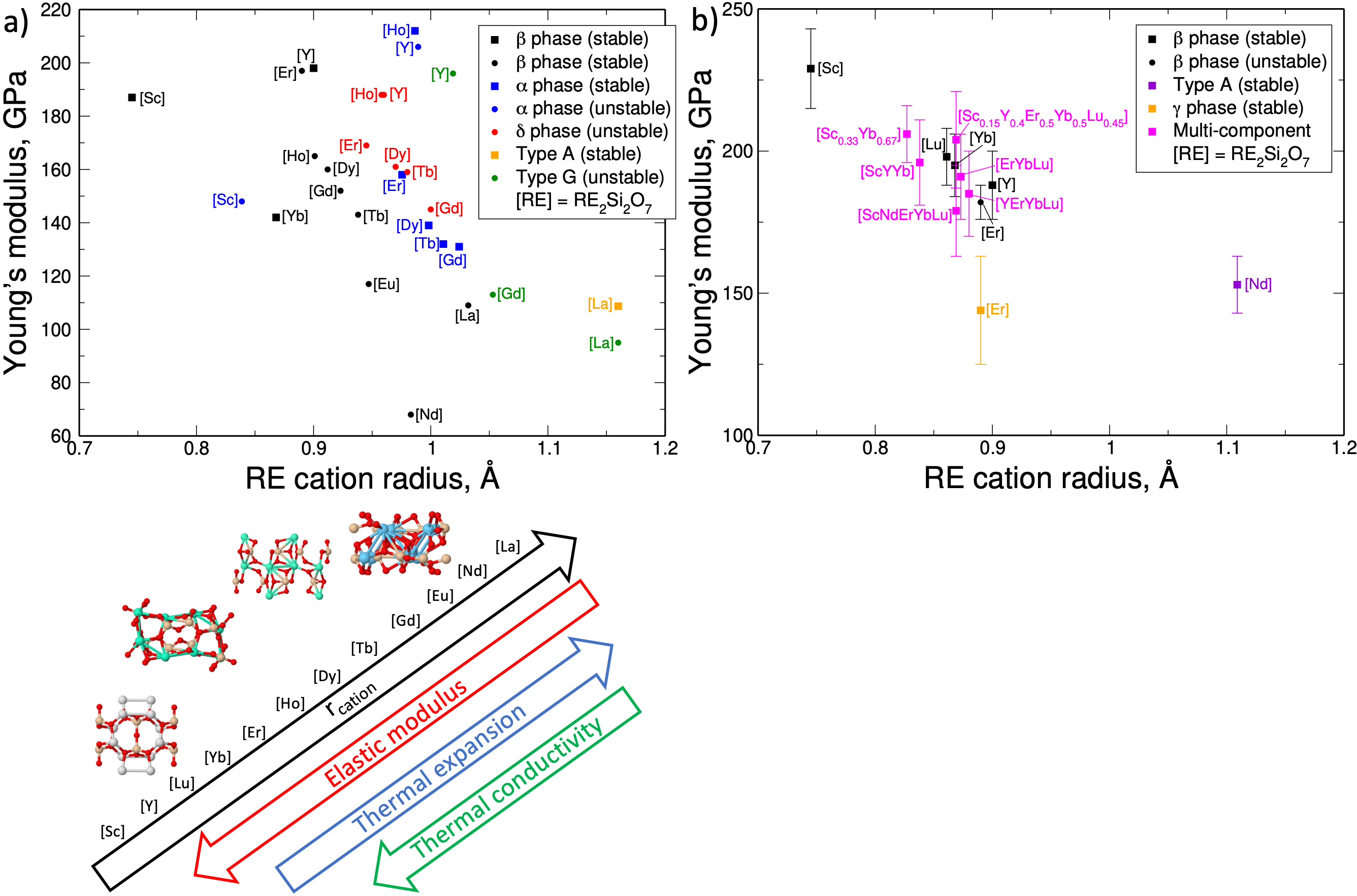
Design rules for the thermal and elastic properties of rare-earth disilicates
6/5/2024 | Elizabeth Opila – University of Virginia
Computational and experimental data were combined to develop design rules for rare-earth disilicates to enable tuning of thermo-mechanical properties to better match the SiC substrate. In the case of Young’s modulus and thermal expansion, these trends also hold for multi-component systems; while the thermal conductivity of multi-component systems is noticeably lower, indicating the potential of such materials to also act as thermal barriers.
Showing 1 to 10 of 40