Self-assembled Peptide-p-electron Supramolular Polymers:Code Sharing and Undergraduate Research

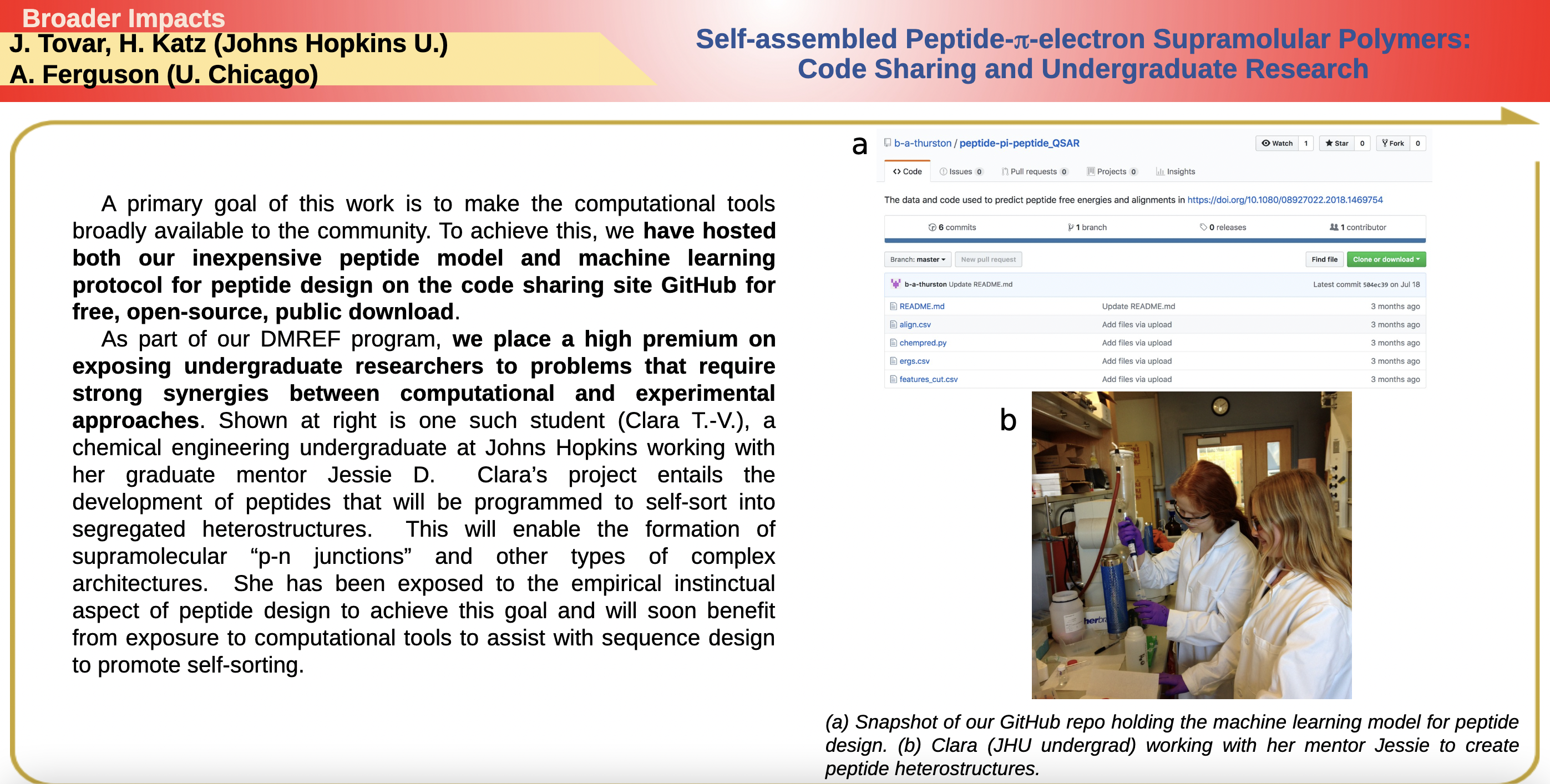
A primary goal of this work is to make the computational tools broadly available to the community. To achieve this, we have hosted both our inexpensive peptide model and machine learning protocol for peptide design on the code sharing site GitHub for free, open-source, public download.
As part of our DMREF program, we place a high premium on exposing undergraduate researchers to problems that require strong synergies between computational and experimental approaches. Shown at right is one such student (Clara T.-V.), a chemical engineering undergraduate at Johns Hopkins working with her graduate mentor Jessie D. Clara’s project entails the development of peptides that will be programmed to self-sort into segregated heterostructures. This will enable the formation of supramolecular “p-n junctions” and other types of complex architectures.
She has been exposed to the empirical instinctual aspect of peptide design to achieve this goal and will soon benefit from exposure to computational tools to assist with sequence design to promote self-sorting.
Snapshot of our GitHub repo holding the machine learning model for peptide design.